Researchers have developed a new way to image the brain with unprecedented resolution and speed. Using this approach, they can locate individual neurons, trace connections between them, and visualize organelles inside neurons, over large volumes of brain tissue.
The new technology combines a method for expanding brain tissue, making it possible to image at higher resolution, with a rapid 3-D microscopy technique known as lattice light-sheet microscopy. In a paper appearing in Science Jan. 17, the researchers showed that they could use these techniques to image the entire fruit fly brain, as well as large sections of the mouse brain, much faster than has previously been possible. The team includes researchers from MIT, the University of California at Berkeley, the Howard Hughes Medical Institute, and Harvard Medical School/Boston Children’s Hospital.
[caption id="attachment_25729" align="alignright" width="640"]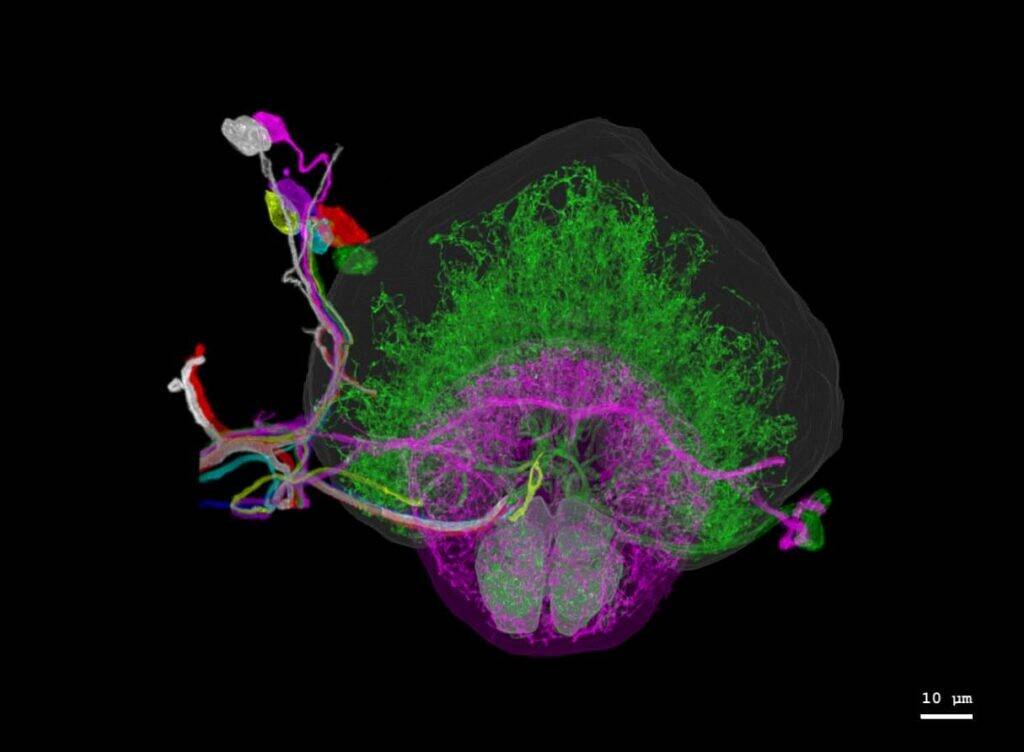 Individually traced dopaminergic neurons in the right hemisphere of a fruit fly brain, innervating the fan-shaped body (green), ellipsoid body (magenta), and noduli (green). Credits Image: Courtesy of the researchers.[/caption]
This technique allows researchers to map large-scale circuits within the brain while also offering unique insight into individual neurons’ functions, says Edward Boyden, the Y. Eva Tan Professor in Neurotechnology, an associate professor of biological engineering and of brain and cognitive sciences at MIT, and a member of MIT’s McGovern Institute for Brain Research, Media Lab, and Koch Institute for Integrative Cancer Research.
“A lot of problems in biology are multiscale,” Boyden says. “Using lattice light-sheet microscopy, along with the expansion microscopy process, we can now image at large scale without losing sight of the nanoscale configuration of biomolecules.”
Boyden is one of the study’s senior authors, along with Eric Betzig, a senior fellow at the Janelia Research Campus and a professor of physics and molecular and cell biology at UC Berkeley. The paper’s lead authors are MIT postdoc Ruixuan Gao, former MIT postdoc Shoh Asano, and Harvard Medical School Assistant Professor Srigokul Upadhyayula.
Individually traced dopaminergic neurons in the right hemisphere of a fruit fly brain, innervating the fan-shaped body (green), ellipsoid body (magenta), and noduli (green). Credits Image: Courtesy of the researchers.[/caption]
This technique allows researchers to map large-scale circuits within the brain while also offering unique insight into individual neurons’ functions, says Edward Boyden, the Y. Eva Tan Professor in Neurotechnology, an associate professor of biological engineering and of brain and cognitive sciences at MIT, and a member of MIT’s McGovern Institute for Brain Research, Media Lab, and Koch Institute for Integrative Cancer Research.
“A lot of problems in biology are multiscale,” Boyden says. “Using lattice light-sheet microscopy, along with the expansion microscopy process, we can now image at large scale without losing sight of the nanoscale configuration of biomolecules.”
Boyden is one of the study’s senior authors, along with Eric Betzig, a senior fellow at the Janelia Research Campus and a professor of physics and molecular and cell biology at UC Berkeley. The paper’s lead authors are MIT postdoc Ruixuan Gao, former MIT postdoc Shoh Asano, and Harvard Medical School Assistant Professor Srigokul Upadhyayula.
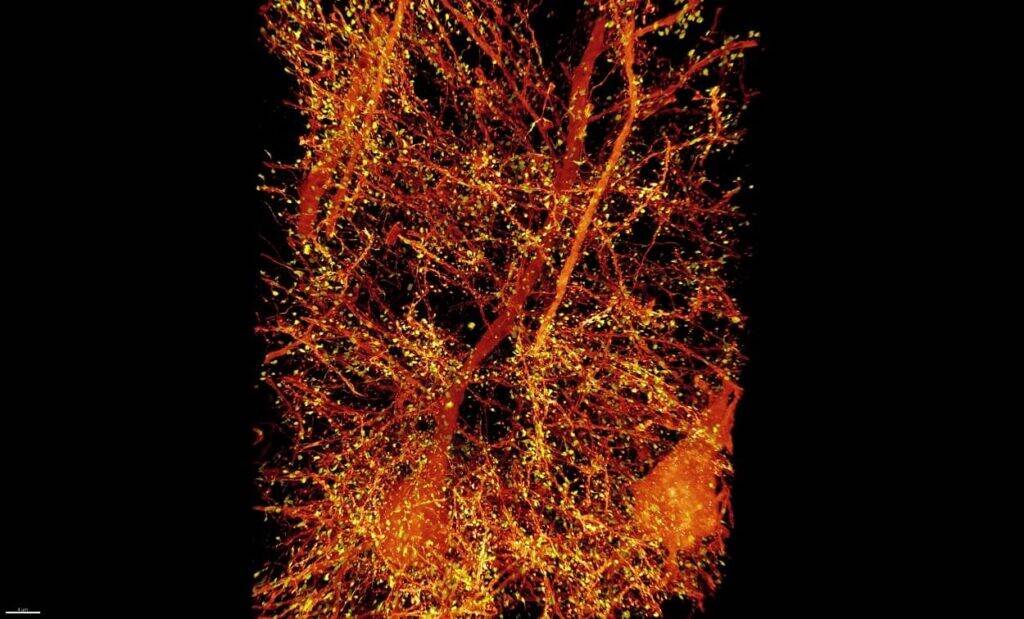 A subset of pyramidal neurons (orange) in the mouse primary somatosensory cortex. The dendritic spines associated with the postsynaptic protein Homer1 are highlighted in yellow. Credits Image: Courtesy of the researchers[/caption]
“The marrying of the lattice light-sheet microscope with expansion microscopy is essential to achieve the sensitivity, resolution, and scalability of the imaging that we’re doing,” Gao says.
Imaging expanded tissue samples generates huge amounts of data — up to tens of terabytes per sample — so the researchers also had to devise highly parallelized computational image-processing techniques that could break down the data into smaller chunks, analyze it, and stitch it back together into a coherent whole.
A subset of pyramidal neurons (orange) in the mouse primary somatosensory cortex. The dendritic spines associated with the postsynaptic protein Homer1 are highlighted in yellow. Credits Image: Courtesy of the researchers[/caption]
“The marrying of the lattice light-sheet microscope with expansion microscopy is essential to achieve the sensitivity, resolution, and scalability of the imaging that we’re doing,” Gao says.
Imaging expanded tissue samples generates huge amounts of data — up to tens of terabytes per sample — so the researchers also had to devise highly parallelized computational image-processing techniques that could break down the data into smaller chunks, analyze it, and stitch it back together into a coherent whole.
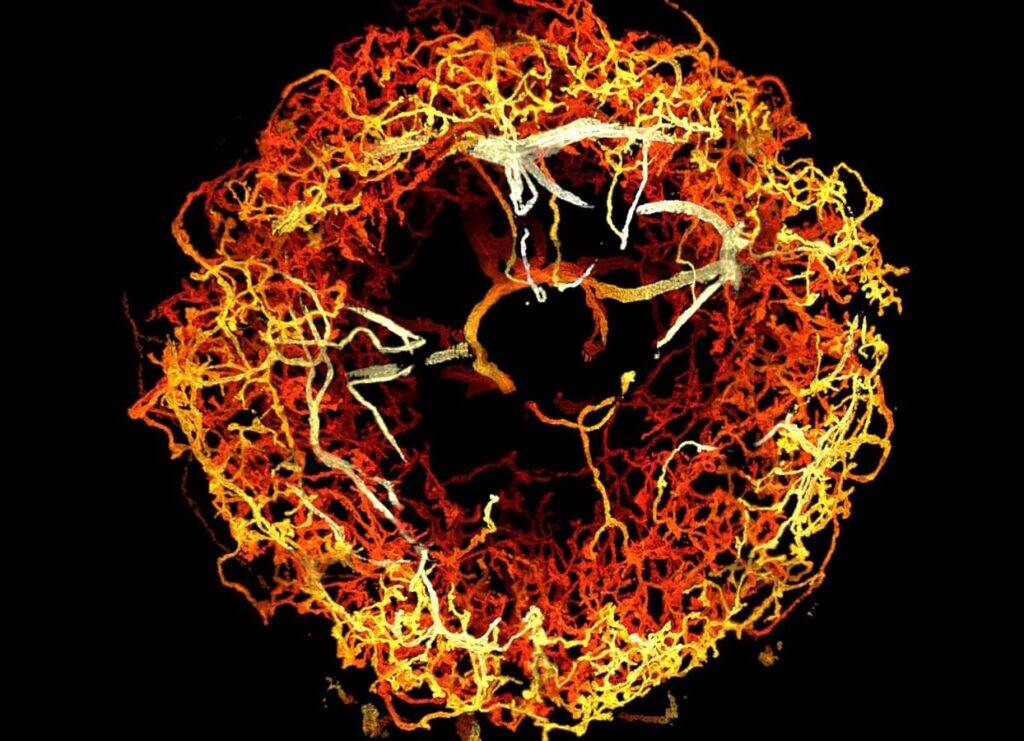 Dopaminergic neurons in the ellipsoid body of a fruit fly brain, color-coded by 3D depth. Credits Image: Courtesy of the researchers.[/caption]
The researchers also studied patterns of axon myelination in different neurons. Myelin is a fatty substance that insulates axons and whose disruption is a hallmark of multiple sclerosis. The researchers were able to compute the thickness of the myelin coating in different segments of axons, and they measured the gaps between stretches of myelin, which are important because they help conduct electrical signals. Previously, this kind of myelin tracing would have required months to years for human annotators to perform.
This technology can also be used to image tiny organelles inside neurons. In the new paper, the researchers identified mitochondria and lysosomes, and they also measured variations in the shapes of these organelles.
Dopaminergic neurons in the ellipsoid body of a fruit fly brain, color-coded by 3D depth. Credits Image: Courtesy of the researchers.[/caption]
The researchers also studied patterns of axon myelination in different neurons. Myelin is a fatty substance that insulates axons and whose disruption is a hallmark of multiple sclerosis. The researchers were able to compute the thickness of the myelin coating in different segments of axons, and they measured the gaps between stretches of myelin, which are important because they help conduct electrical signals. Previously, this kind of myelin tracing would have required months to years for human annotators to perform.
This technology can also be used to image tiny organelles inside neurons. In the new paper, the researchers identified mitochondria and lysosomes, and they also measured variations in the shapes of these organelles.
 Individually traced dopaminergic neurons in the right hemisphere of a fruit fly brain, innervating the fan-shaped body (green), ellipsoid body (magenta), and noduli (green). Credits Image: Courtesy of the researchers.[/caption]
This technique allows researchers to map large-scale circuits within the brain while also offering unique insight into individual neurons’ functions, says Edward Boyden, the Y. Eva Tan Professor in Neurotechnology, an associate professor of biological engineering and of brain and cognitive sciences at MIT, and a member of MIT’s McGovern Institute for Brain Research, Media Lab, and Koch Institute for Integrative Cancer Research.
“A lot of problems in biology are multiscale,” Boyden says. “Using lattice light-sheet microscopy, along with the expansion microscopy process, we can now image at large scale without losing sight of the nanoscale configuration of biomolecules.”
Boyden is one of the study’s senior authors, along with Eric Betzig, a senior fellow at the Janelia Research Campus and a professor of physics and molecular and cell biology at UC Berkeley. The paper’s lead authors are MIT postdoc Ruixuan Gao, former MIT postdoc Shoh Asano, and Harvard Medical School Assistant Professor Srigokul Upadhyayula.
Individually traced dopaminergic neurons in the right hemisphere of a fruit fly brain, innervating the fan-shaped body (green), ellipsoid body (magenta), and noduli (green). Credits Image: Courtesy of the researchers.[/caption]
This technique allows researchers to map large-scale circuits within the brain while also offering unique insight into individual neurons’ functions, says Edward Boyden, the Y. Eva Tan Professor in Neurotechnology, an associate professor of biological engineering and of brain and cognitive sciences at MIT, and a member of MIT’s McGovern Institute for Brain Research, Media Lab, and Koch Institute for Integrative Cancer Research.
“A lot of problems in biology are multiscale,” Boyden says. “Using lattice light-sheet microscopy, along with the expansion microscopy process, we can now image at large scale without losing sight of the nanoscale configuration of biomolecules.”
Boyden is one of the study’s senior authors, along with Eric Betzig, a senior fellow at the Janelia Research Campus and a professor of physics and molecular and cell biology at UC Berkeley. The paper’s lead authors are MIT postdoc Ruixuan Gao, former MIT postdoc Shoh Asano, and Harvard Medical School Assistant Professor Srigokul Upadhyayula.
Large-scale imaging
In 2015, Boyden’s lab developed a way to generate very high-resolution images of brain tissue using an ordinary light microscope. Their technique relies on expanding tissue before imaging it, allowing them to image the tissue at a resolution of about 60 nanometers. Previously, this kind of imaging could be achieved only with very expensive high-resolution microscopes, known as super-resolution microscopes. In the new study, Boyden teamed up with Betzig and his colleagues at HHMI’s Janelia Research Campus to combine expansion microscopy with lattice light-sheet microscopy. This technology, which Betzig developed several years ago, has some key traits that make it ideal to pair with expansion microscopy: It can image large samples rapidly, and it induces much less photodamage than other fluorescent microscopy techniques. [caption id="attachment_25730" align="alignright" width="640"] A subset of pyramidal neurons (orange) in the mouse primary somatosensory cortex. The dendritic spines associated with the postsynaptic protein Homer1 are highlighted in yellow. Credits Image: Courtesy of the researchers[/caption]
“The marrying of the lattice light-sheet microscope with expansion microscopy is essential to achieve the sensitivity, resolution, and scalability of the imaging that we’re doing,” Gao says.
Imaging expanded tissue samples generates huge amounts of data — up to tens of terabytes per sample — so the researchers also had to devise highly parallelized computational image-processing techniques that could break down the data into smaller chunks, analyze it, and stitch it back together into a coherent whole.
A subset of pyramidal neurons (orange) in the mouse primary somatosensory cortex. The dendritic spines associated with the postsynaptic protein Homer1 are highlighted in yellow. Credits Image: Courtesy of the researchers[/caption]
“The marrying of the lattice light-sheet microscope with expansion microscopy is essential to achieve the sensitivity, resolution, and scalability of the imaging that we’re doing,” Gao says.
Imaging expanded tissue samples generates huge amounts of data — up to tens of terabytes per sample — so the researchers also had to devise highly parallelized computational image-processing techniques that could break down the data into smaller chunks, analyze it, and stitch it back together into a coherent whole.
Using this technique, it is possible to analyze millions of synapses in just a few daysIn the Science paper, the researchers demonstrated the power of their new technique by imaging layers of neurons in the somatosensory cortex of mice, after expanding the tissue volume fourfold. They focused on a type of neuron known as pyramidal cells, one of the most common excitatory neurons found in the nervous system. To locate synapses, or connections, between these neurons, they labeled proteins found in the presynaptic and postsynaptic regions of the cells. This also allowed them to compare the density of synapses in different parts of the cortex. Using this technique, it is possible to analyze millions of synapses in just a few days. “We counted clusters of postsynaptic markers across the cortex, and we saw differences in synaptic density in different layers of the cortex,” Gao says. “Using electron microscopy, this would have taken years to complete.” [caption id="attachment_25731" align="alignright" width="640"]
 Dopaminergic neurons in the ellipsoid body of a fruit fly brain, color-coded by 3D depth. Credits Image: Courtesy of the researchers.[/caption]
The researchers also studied patterns of axon myelination in different neurons. Myelin is a fatty substance that insulates axons and whose disruption is a hallmark of multiple sclerosis. The researchers were able to compute the thickness of the myelin coating in different segments of axons, and they measured the gaps between stretches of myelin, which are important because they help conduct electrical signals. Previously, this kind of myelin tracing would have required months to years for human annotators to perform.
This technology can also be used to image tiny organelles inside neurons. In the new paper, the researchers identified mitochondria and lysosomes, and they also measured variations in the shapes of these organelles.
Dopaminergic neurons in the ellipsoid body of a fruit fly brain, color-coded by 3D depth. Credits Image: Courtesy of the researchers.[/caption]
The researchers also studied patterns of axon myelination in different neurons. Myelin is a fatty substance that insulates axons and whose disruption is a hallmark of multiple sclerosis. The researchers were able to compute the thickness of the myelin coating in different segments of axons, and they measured the gaps between stretches of myelin, which are important because they help conduct electrical signals. Previously, this kind of myelin tracing would have required months to years for human annotators to perform.
This technology can also be used to image tiny organelles inside neurons. In the new paper, the researchers identified mitochondria and lysosomes, and they also measured variations in the shapes of these organelles.